Discuit includes a library of predefined, mostly epidemiological, models. Models can also be user defined, or generated using one of the predefined models as a template. This article focuses on the latter approach, describing the model generating function with an overview of default components.
Generating models
Pre defined models, such as the SIS example given in the paper, can be invoked by calling:
model = GenerateModel("SIS", c(100, 1))## Julia version 1.0.0 at location /home/martin/dev/julia/julia-1.0.0-linux-x86_64/julia-1.0.0/bin will be used.## Loading setup script for JuliaCall...## Finish loading setup script for JuliaCall.The modelName and initialCondition are required to call GenerateModel. The default observation model is Gaussian and an optional third parameter can be used to specify the observation error \(\sigma_{ERR}\):
model = GenerateModel("SIS", c(100, 1), 2)The following gives an overview of important defaults that the user should note before proceeding with their analysis.
Standard Kermack-McKendrick models
The standard Kermack-McKendrick SIR model can be used to model diseases which confer lasting immunity or situations where infected individuals have been detected and quarantined for treatment (Kermack and McKendrick, 1991). ADD REF: kermack_contributions_1991
Its variations include the susceptible-infectious ("SI") model, a very basic model with only one type of event. Individuals who become infected remain infected for the duration of trajectory.

The classic Kermack-McKendrick susceptible-infectious-recovered ("SIR") model includes an extra ‘recovery’ event and an additional compartment for recovered individuals. The susceptible-infectious-susceptible ("SIS") model is an extension of the classic Kermack-McKendrick SIR model for diseases which do not confer lasting immunity:
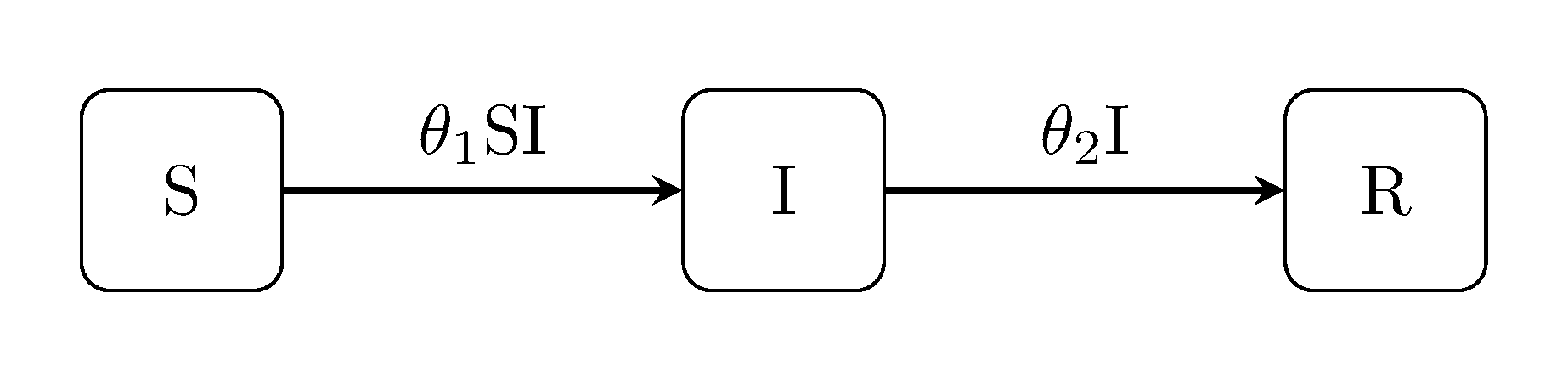

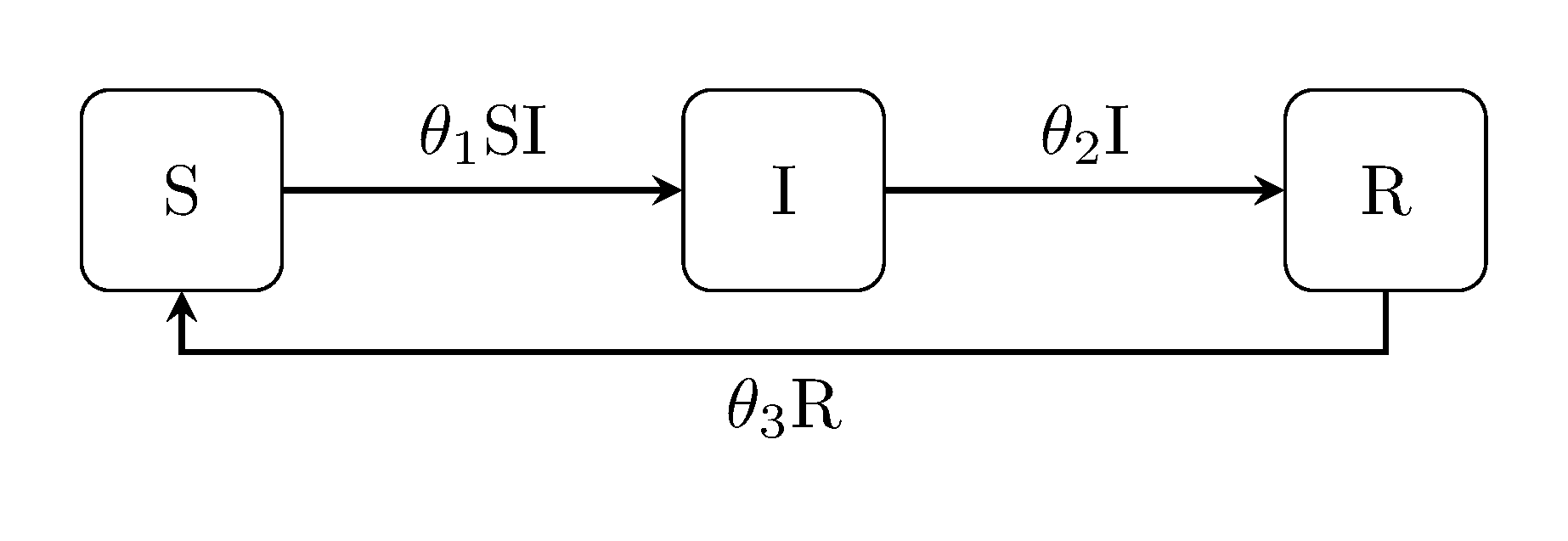
The susceptible-infectious-recovered-susceptible ("SIRS") model incorporates all of the above, i.e. it is for diseases which do not confer long lasting immunity:
Latent Kermack-McKendrick models
The next class of models extend the classic Kermack-McKendrick by accounting for an exposed state E between infection and the onset of infectiousness. For example, the susceptible-exposed-infectious SEI model:
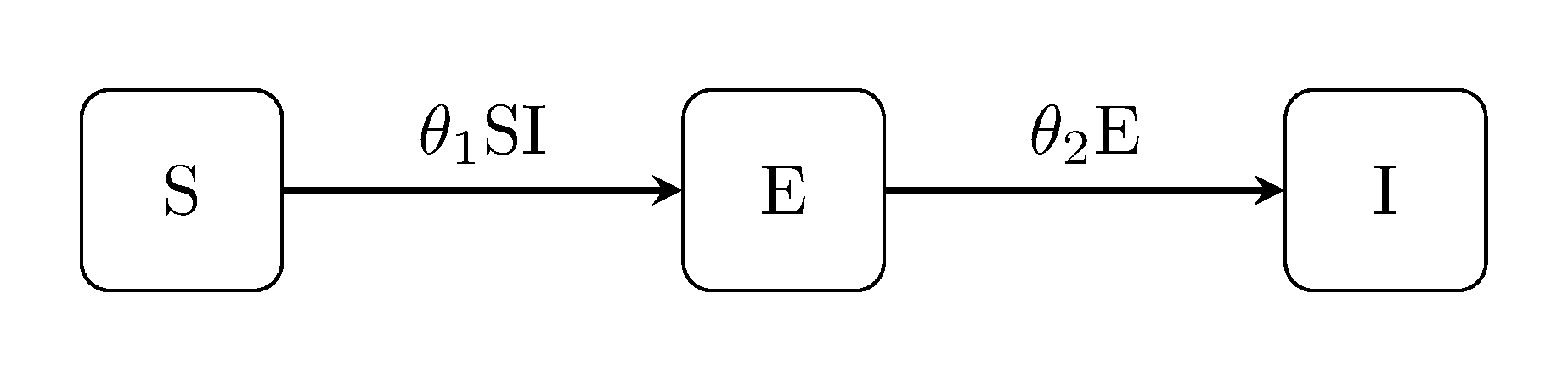
The latent Kermack-McKendrick models can be extended in the same way, i.e, the SEIR model, SEIS and SEIRS:
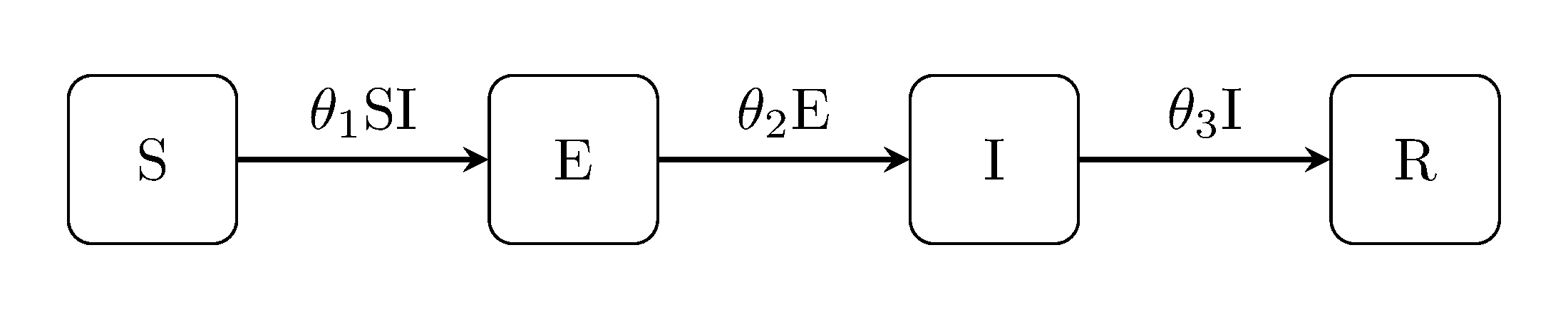
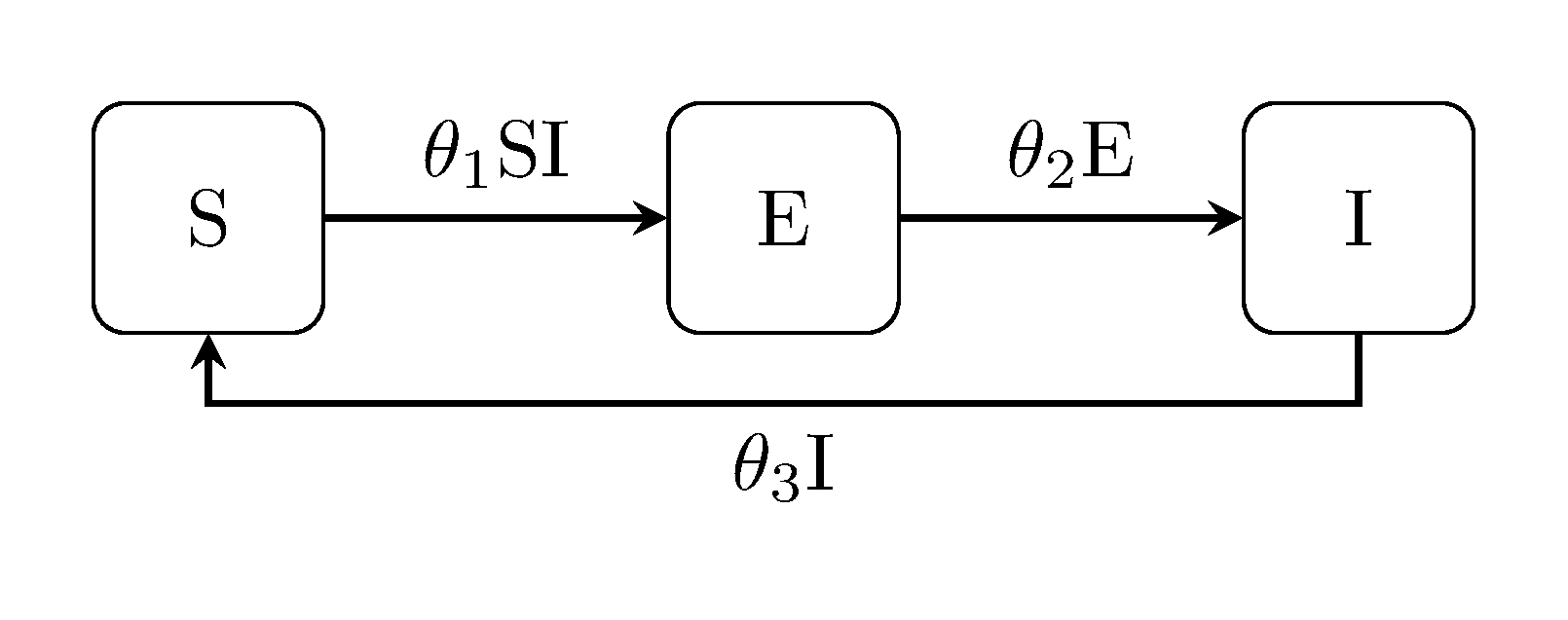

Example
Using the generated model is easy, for example:
parameters = c(0.003, 0.1)
xi = RunSimulation(model, parameters)
PlotTrajectory(xi$trajectory)